PennCNV
#
Find similar titles
- (rev. 3)
- Hyungyong Kim
Structured data
- About
- SNP array
- Copy-number variation
- Programming Language
- Perl
- URL
- http://www.openbioinformatics.org/penncnv/
SNP array 데이터에서 CNV를 탐지하는 프로그램
PennCNV implements a HMM that integrates multiple sources of information to infer CNV calls for individual genotyped samples. It differs form segmentation-based algorithm in that it considered SNP allelic ratio distribution as well as other factors, in addition to signal intensity alone.
In addition, PennCNV can optionally utilize family information to generate family-based CNV calls by several different algorithms. Furthermore, PennCNV can generate CNV calls given a specific set of candidate CNV regions, through a validation-calling algorithm.
Table of Contents
PennCNV-Affy #
Data processing protocol for Genome-Wide Human SNP Array 6.0. It converts raw CEL files into a signal intensity file that contains Log R Ratio (LRR) and B Allele Frequency (BAF) vlaues.
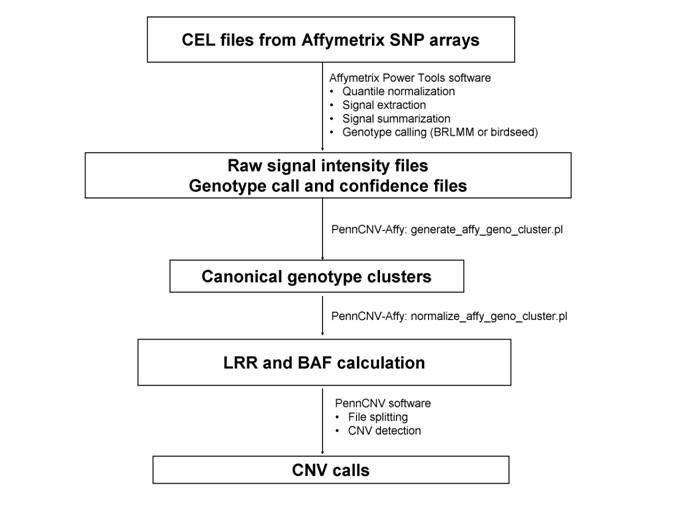
Procedure
- Generate genotyping calls from CEL file using Affymetrix Power Tools apt-probeset-genotype program
- Allele-specific signal extraction from CEL files using Affymetrix Power Tools apt-probeset-summarize
- Generate canonical genotype clustering file by generate_affy_geno_cluster.pl
- LRR and BAF calculation by normalize_affy_geno_cluster.pl
- Split the signal file into individual files and running CNV calling detect_cnv.pl
CNV calling #
Input file from
- PennCNV-Affy
- Illumina Report
- BeadStudio project file
Overview
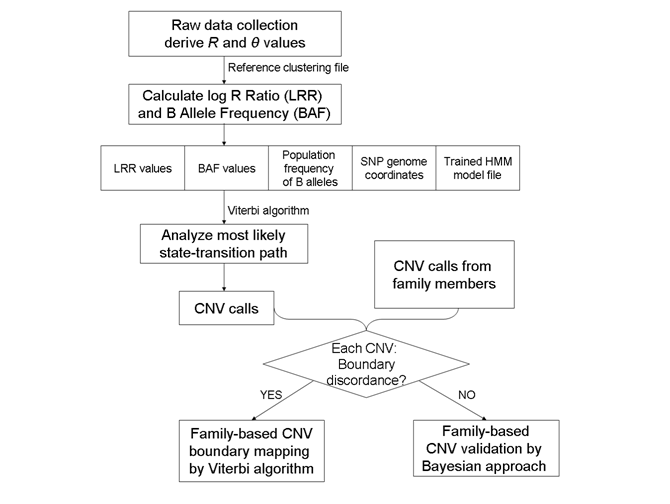
Output example (rawcnv)
chr1:160659106-160915937 numsnp=79 length=256,832 state2,cn=1 gw6.Genomewide6 startsnp=CN_442866 endsnp=CN_442942
chr1:163703204-165581459 numsnp=645 length=1,878,256 state2,cn=1 gw6.Genomewide6 startsnp=CN_453837 endsnp=CN_439198
chr1:165901010-169387961 numsnp=1191 length=3,486,952 state2,cn=1 gw6.Genomewide6 startsnp=CN_440397 endsnp=CN_452581
chr1:169448874-169643388 numsnp=53 length=194,515 state2,cn=1 gw6.Genomewide6 startsnp=CN_452606 endsnp=CN_452661
chr1:169825272-171629811 numsnp=615 length=1,804,540 state2,cn=1 gw6.Genomewide6 startsnp=CN_453958 endsnp=CN_439314
chr2:7876421-8183989 numsnp=120 length=307,569 state2,cn=1 gw6.Genomewide6 startsnp=CN_202764 endsnp=CN_860524
chr2:54108877-55094559 numsnp=360 length=985,683 state2,cn=1 gw6.Genomewide6 startsnp=CN_846479 endsnp=CN_853178
관련자료 #
논문
- PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data Genome Research
Incoming Links #
Related Products (Product 0) #
Related Articles (Article 1) #
Suggested Pages #
- 0.079 EquineSNP50 BeadChip
- 0.070 Genetic variation
- 0.063 CanineHD Whole-Genome Genotyping BeadChip
- 0.053 Prostate cancer
- 0.052 saasCNV
- 0.051 Identification of cancer-driver genes in focal genomic alterations from whole genome sequencing data
- 0.045 Amplicon
- 0.040 Cattle
- 0.040 Gene expression
- 0.038 QTL
- More suggestions...